
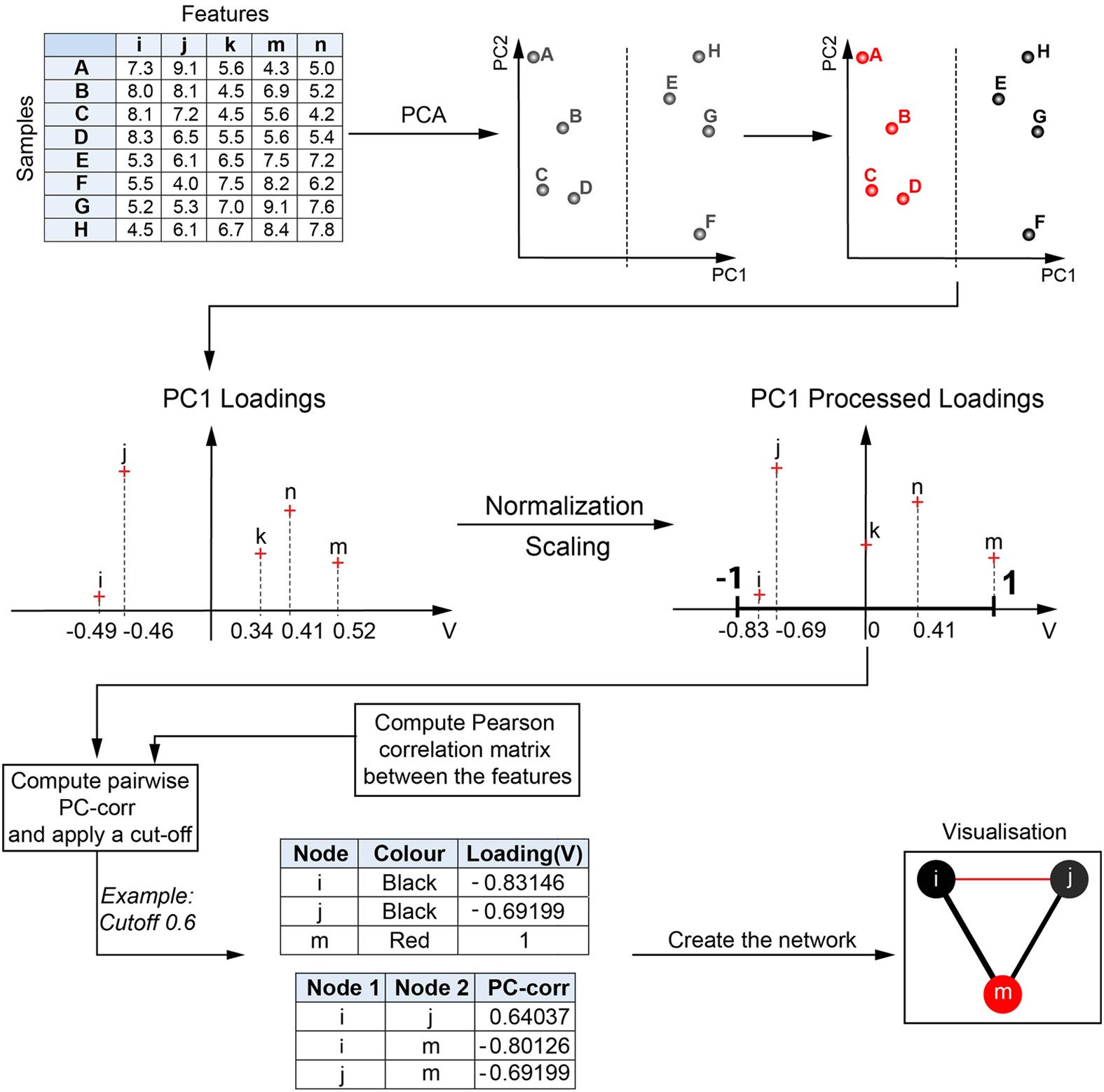
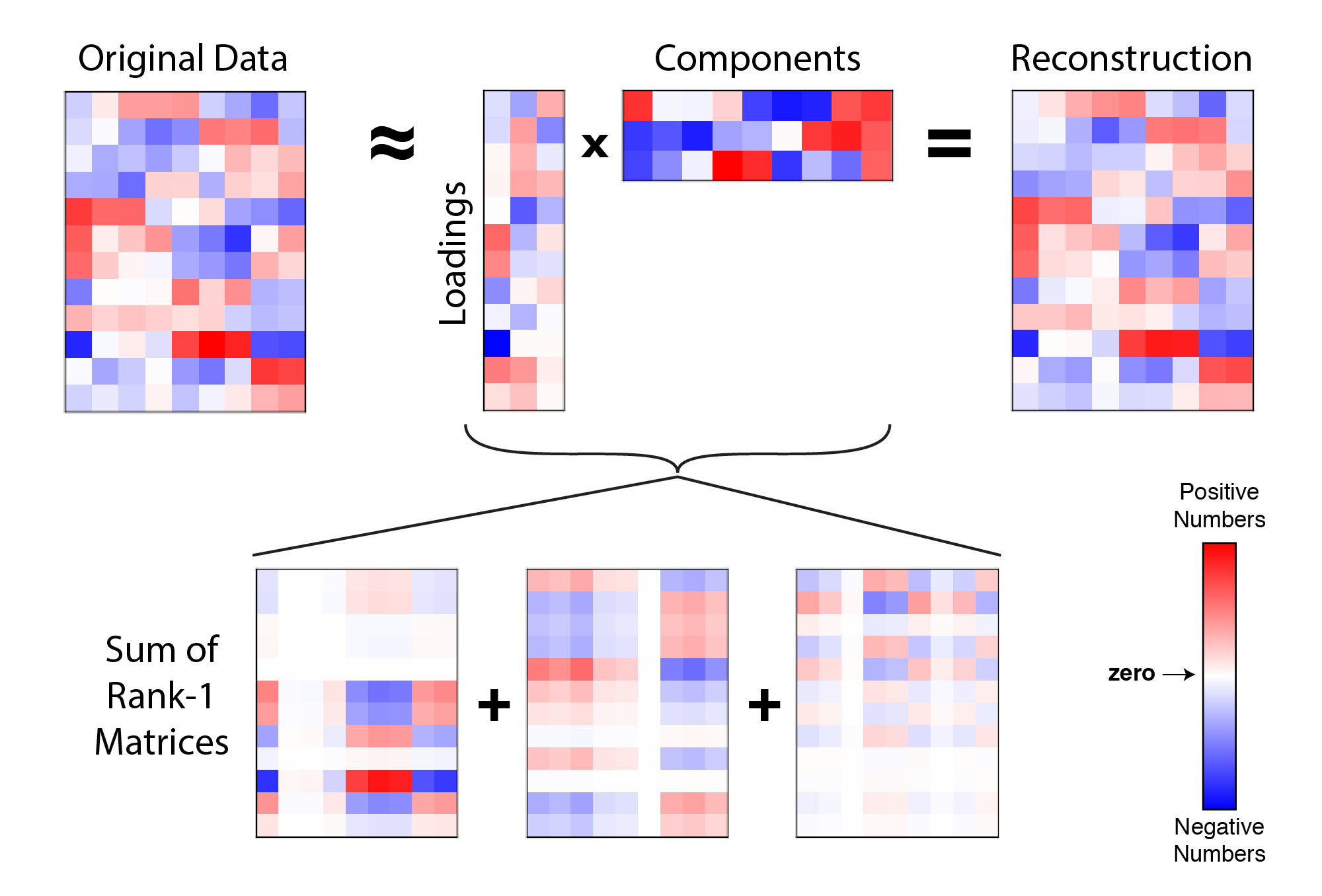
At any rate, I guarantee you can master PCA without fully understanding the process. SVD-based PCA does not tolerate missing values (but there are solutions we will cover shortly)įor a more elaborate explanation with introductory linear algebra, here is an excellent free SVD tutorial I found online.The columns of should be mean-centered, so that the covariance matrix.PCs are ordered by the decreasing amount of variance explained.Consequently, multiplying all scores and loadings recovers. The loading factors of the PC are directly given in the row in. The scores from the first PCs result from multiplying the first columns of with the upper-left submatrix of. PCA reduces the dimensions of your data set down to principal components (PCs). The key difference of SVD compared to a matrix diagonalization ( ) is that and are distinct orthonormal (orthogonal and unit-vector) matrices. These matrices are of size, and, respectively. Where is the matrix with the eigenvectors of, is the diagonal matrix with the singular values and is the matrix with the eigenvectors of. One of the most popular methods is the singular value decomposition (SVD). The SVD algorithm breaks down a matrix of size into three pieces,
There are numerous PCA formulations in the literature dating back as long as one century, but all in all PCA is pure linear algebra. Notwithstanding the focus on life sciences, it should still be clear to others than biologists. In case PCA is entirely new to you, there is an excellent Primer from Nature Biotechnology that I highly recommend. The PLS is worth an entire post and so I will refrain from casting a second spotlight. Its counterpart, the partial least squares (PLS), is a supervised method and will perform the same sort of covariance decomposition, albeit building a user-defined number of components (frequently designated as latent variables) that minimize the SSE from predicting a specified outcome with an ordinary least squares (OLS). It is an unsupervised method, meaning it will always look into the greatest sources of variation regardless of the data structure. PCA is particularly powerful in dealing with multicollinearity and variables that outnumber the samples ( ). From the detection of outliers to predictive modeling, PCA has the ability of projecting the observations described by variables into few orthogonal components defined at where the data ‘stretch’ the most, rendering a simplified overview. Principal component analysis (PCA) is routinely employed on a wide range of problems.


 0 kommentar(er)
0 kommentar(er)
